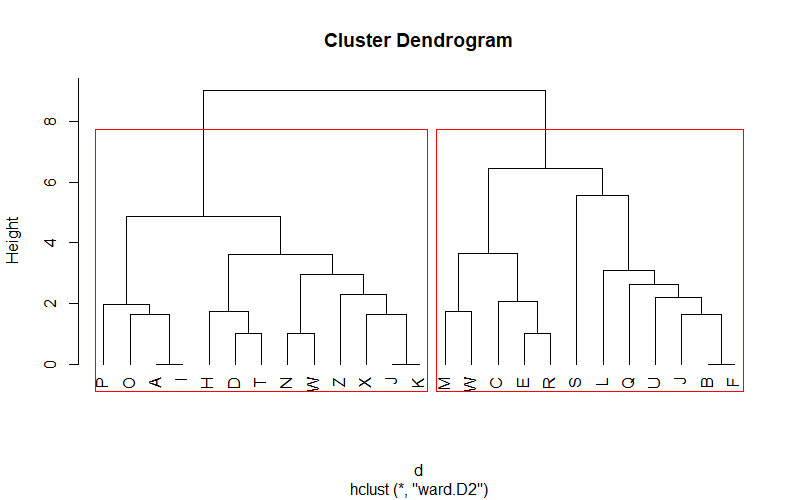
クラスター分析とデンドログラムの表示
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 | library(readr)library(dplyr)library(magrittr)library(reshape2)library(ggplot2)library(fpc)# データの概要 label item1 item2 item3 item4 item5 item6 item7 item8 item9 item101 A 5 5 5 5 5 5 5 5 5 52 B 5 4 4 4 4 4 4 4 3 33 C 5 5 5 5 5 5 5 4 5 14 D 5 5 5 5 5 5 5 3 4 36 E 5 5 5 5 5 5 5 5 3 17 F 5 4 4 4 4 4 4 4 3 3# Euclidean Distanced <- dist(dat[, -1], method = "euclidean")# Hierarchical Clustering with Ward Methodcluster <- hclust(d, method = "ward.D2")# Dendrogramplot(cluster, labels = dat$label, hang = -1)# k = nのクラスター数で囲いを入れるrect.hclust(cluster, k = 2, border = "red") |
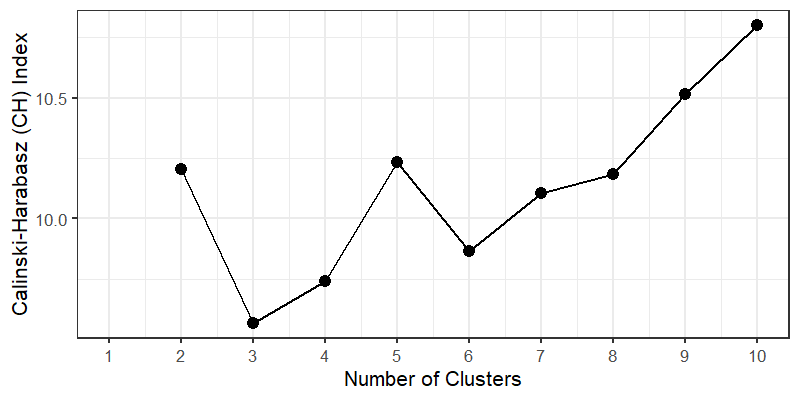
クラスター数の決定
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 | ##Calinski-Harabasz (CH) index(Pseudo F)# total sum of squarestotss <- function(dmatrix){ grandmean <- apply(dmatrix, 2, FUN = mean) sum(apply(dmatrix, 1, FUN = function(row){sqr_edist(row, grandmean)}))}# wssPseudo_F <- function(dmatrix, kmax){ npts <- dim(dmatrix)[1] # number of rows. totss <- totss(dmatrix) wss <- numeric(kmax) crit <- numeric(kmax) wss[1] <- (npts - 1) * sum(apply(dmatrix, 2, var)) for(k in 2 : kmax){ d <- dist(dmatrix, method = "euclidean") pfit <- hclust(d, method = "ward.D2") labels <- cutree(pfit, k = k) wss[k] <- wss.total(dmatrix, labels) } bss <- totss - wss # bssの計算 crit.num <- bss / (0 : (kmax - 1)) # bss/k-1 crit.denom <- wss/(npts - 1 : kmax) # wss/n-k list(crit = crit.num / crit.denom, wss = wss, totss = totss, test= crit.num) # crit = (bss/k-1) / (wss/n-k)}# 結果の可視化(CH indexが極大となるクラスター数を採用)Fcrit <- Pseudo_F(dat[, -1], 10)Fframe <- data.frame(k = 1 : 10, ch = Fcrit$crit)Fframe <- melt(Fframe, id.vars = c("k"), variable.name = "measure", value.name = "score")ggplot(Fframe, aes(x = k, y = score, color = measure)) + geom_point(aes(shape = measure), color = "black", size = 6, shape = 20) + geom_line(aes(linetype = measure), color = "black", size = 1) + theme_bw(base_size = 16, base_family = '') + scale_x_continuous(breaks = 1 : 10, labels = 1 : 10) + labs(x = "Number of Clusters", y = "Calinski-Harabasz (CH) Index") + theme(legend.position = 'none')# クラスタの安定度kbest.p <- 4 # 任意の数cboot.hclust <- clusterboot(dat[, -1], clustermethod = hclustCBI, method = "ward.D2", k = kbest.p)bootMean.data <- data.frame(cluster = 1 : kbest.p, bootMeans = cboot.hclust$bootmean)round(bootMean.data, digit = 2) |